Genomic DNA sequences are being determined at an increasingly rapid pace. This has created a need for more efficient techniques to determine which parts of these sequences are bound in-vivo by the proteins controlling processes; such as gene expression, DNA replication and chromosomal mechanics.
A whole-genome approach was established to identify and characterize such DNA sequences. The method of chromatin immunoprecipitation, combined with microarrays (ChIP-Chip), is a powerful tool for genome-wide analysis of protein binding. It has also become a widely-used method for genome-wide localization of protein-DNA interactions.
The first step in the ChIP-Chip procedure is to fix protein-DNA interactions in living cells by chemical crosslinking. The crosslinker must be small to diffuse fast into the cells. In practice, formaldehyde is used in most ChIP-Chip experiments. After cell lysis, the DNA is fragmented by sonication. This extract is then subjected to immunoprecipitation (IP) with a specific antibody against the protein of interest.
DNA bound by the protein will be coprecipitated and enriched, compared to DNA not bound by the respective protein. To facilitate immunoprecipitation and subsequent washing, antibodies are usually coupled to either agarose- or magnetic beads via protein A or G. After reversion of crosslinking, the DNA is purified by phenol extraction or commercial polymerase chain reaction (PCR) cleanup kits.
Often, an amplification step is included after DNA purification. Two different fluorescence labels are used to label the IP DNA, and a hybridization-control DNA, respectively. Usually, total DNA before IP (input DNA) is used as hybridization control.
The two differentially-labeled DNAs are hybridized to the same microarray and the difference in fluorescence intensity gives a measure of the enrichment .
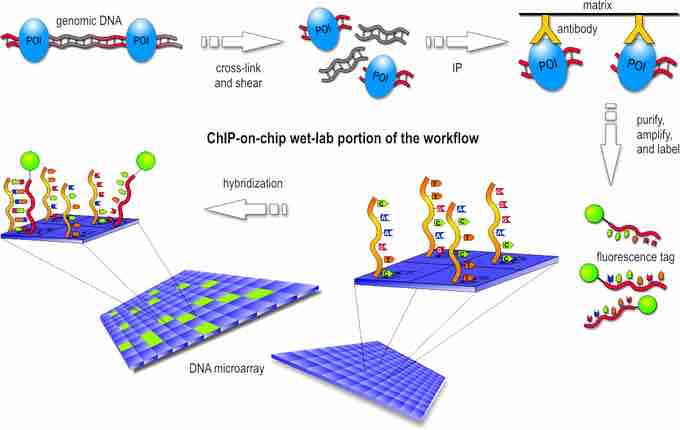
ChIP-chip procedure
ChIP-chip workflow performed in laboratory