Deltaproteobacteria is a class of Proteobacteria . All species of this group are, like all Proteobacteria, Gram-negative.
Deltaproteobacteria
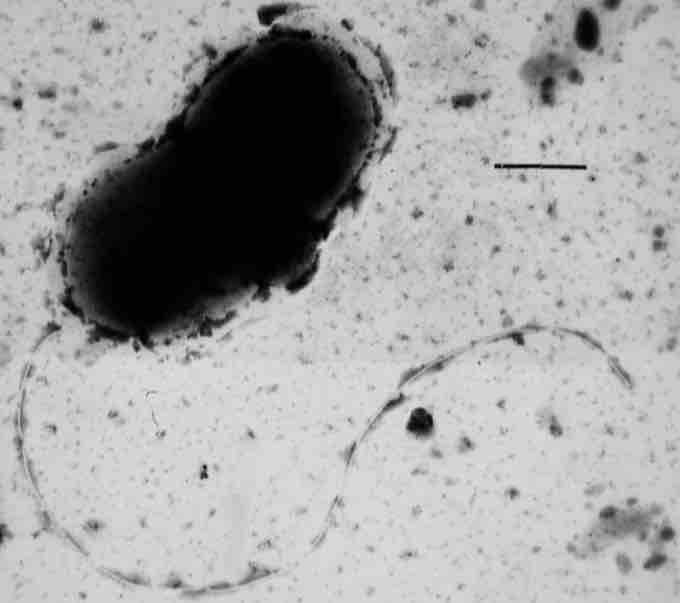
Desulfovibrio vulgaris
The Deltaproteobacteria comprise a branch of predominantly aerobic genera, the fruiting-body-forming Myxobacteria that release myxospores in unfavorable environments. It is a branch of strictly anaerobic genera, which contains most of the known sulfate- (Desulfovibrio, Desulfobacter, Desulfococcus, Desulfonema, etc. ) and sulfur-reducing bacteria (e.g. Desulfuromonas spp.) alongside several other anaerobic bacteria with different physiology (e.g. ferric iron-reducing Geobacter spp. and syntrophic Pelobacter and Syntrophus spp.). A pathogenic intracellular Deltaproteobacteria has recently been identified.
The myxobacteria ("slime bacteria") are a group of bacteria that predominantly live in the soil and feed on insoluble organic substances. The myxobacteria have very large genomes, relative to other bacteria, e.g. 9–10 million nucleotides. Sorangium cellulosum has the largest known (as of 2008) bacterial genome, at 13.0 million nucleotides. Myxobacteria are included among the delta group of proteobacteria, a large taxon of Gram-negative forms.
Myxobacteria can move actively by gliding. They typically travel in swarms (also known as wolf packs), containing many cells kept together by intercellular molecular signals. Individuals benefit from aggregation as it allows accumulation of extracellular enzymes which are used to digest food. This in turn increases feeding efficiency. Myxobacteria produce a number of biomedically and industrially useful chemicals, such as antibiotics. They export those chemicals outside of the cell.
The currently accepted taxonomy is based on the List of Prokaryotic Names with Standing in Nomenclature (LPSN) and National Center for Biotechnology Information (NCBI) and the phylogeny is based on 16S rRNA-based LTP release 106 by 'The All-Species Living Tree' Project.